3.1 AlphaFold: a solution to a 50-year-old grand challenge in biology
Proteins are the building blocks of life, how they work and what they do is determined by its 3-D shape, structure is function. Proteins consist out of amino acid strings. Since 1994 the world wide experiment Critical Assessment of protein Structure Prediction, or CASP is held biennial. In 2018 Deepmind won at its first entry and was was head-and-shoulders above other teams in 2020.
This will change medicine. It will change research. It will change bioengineering. It will change everything
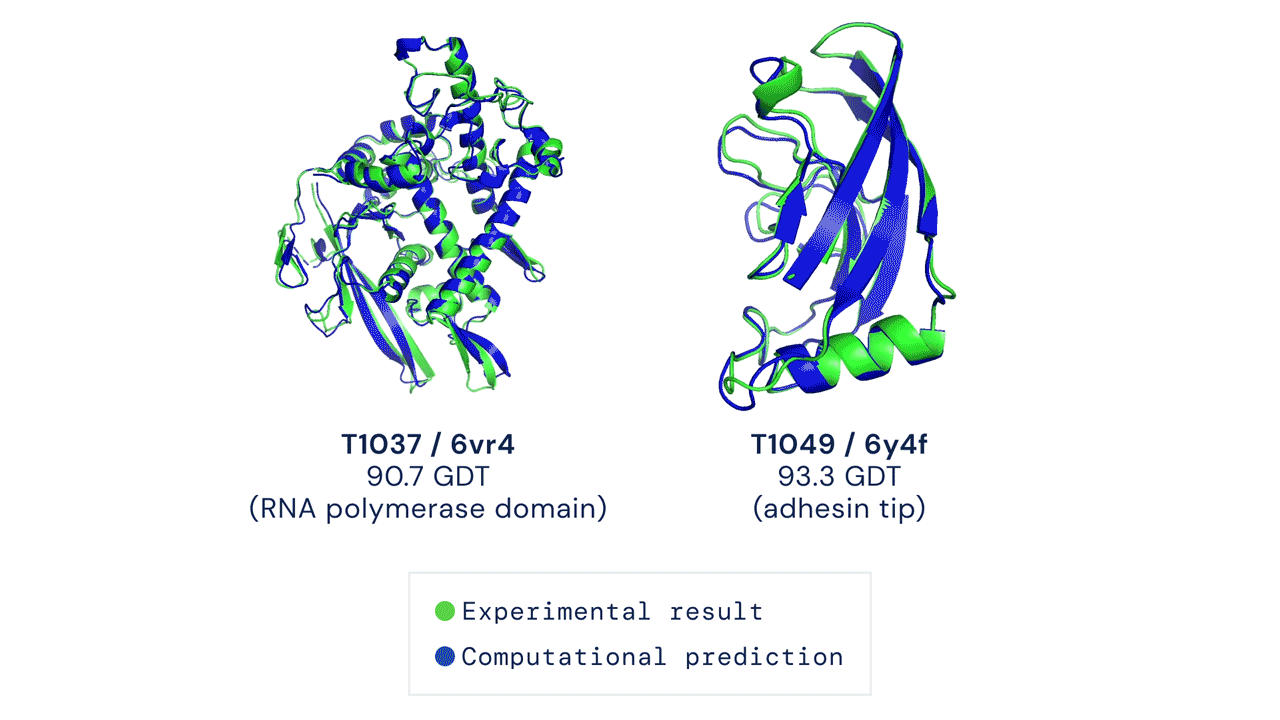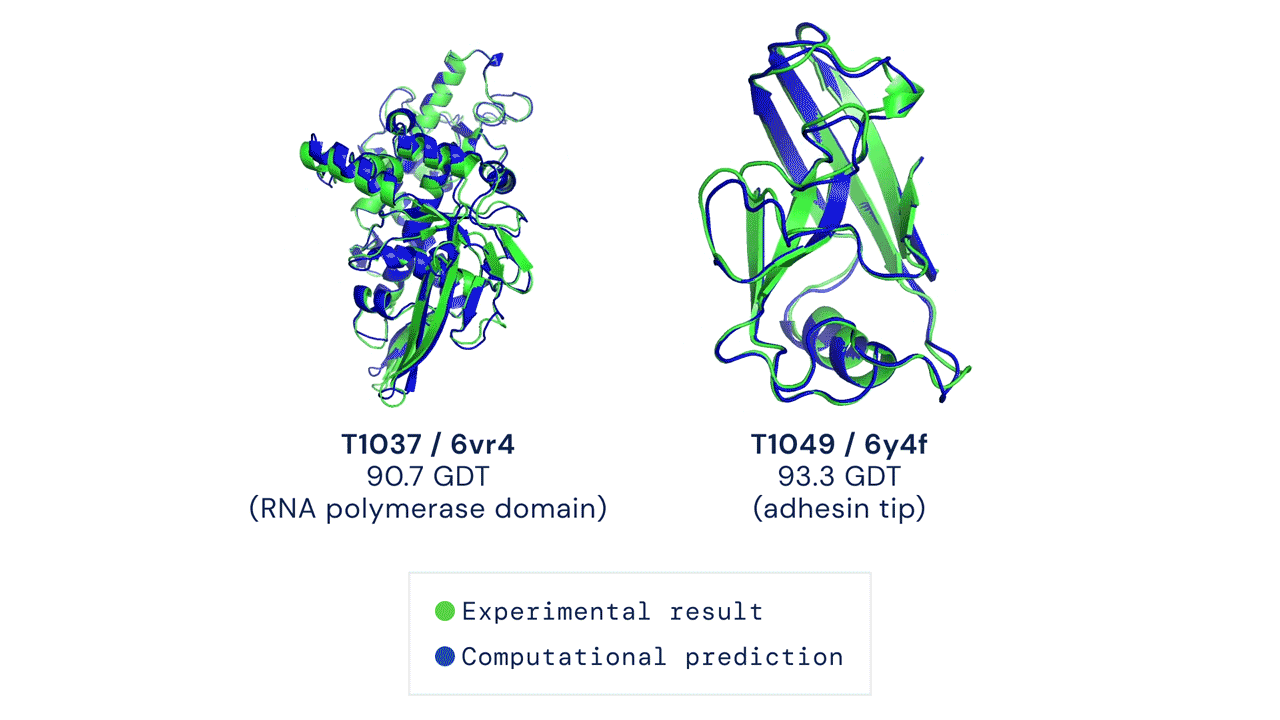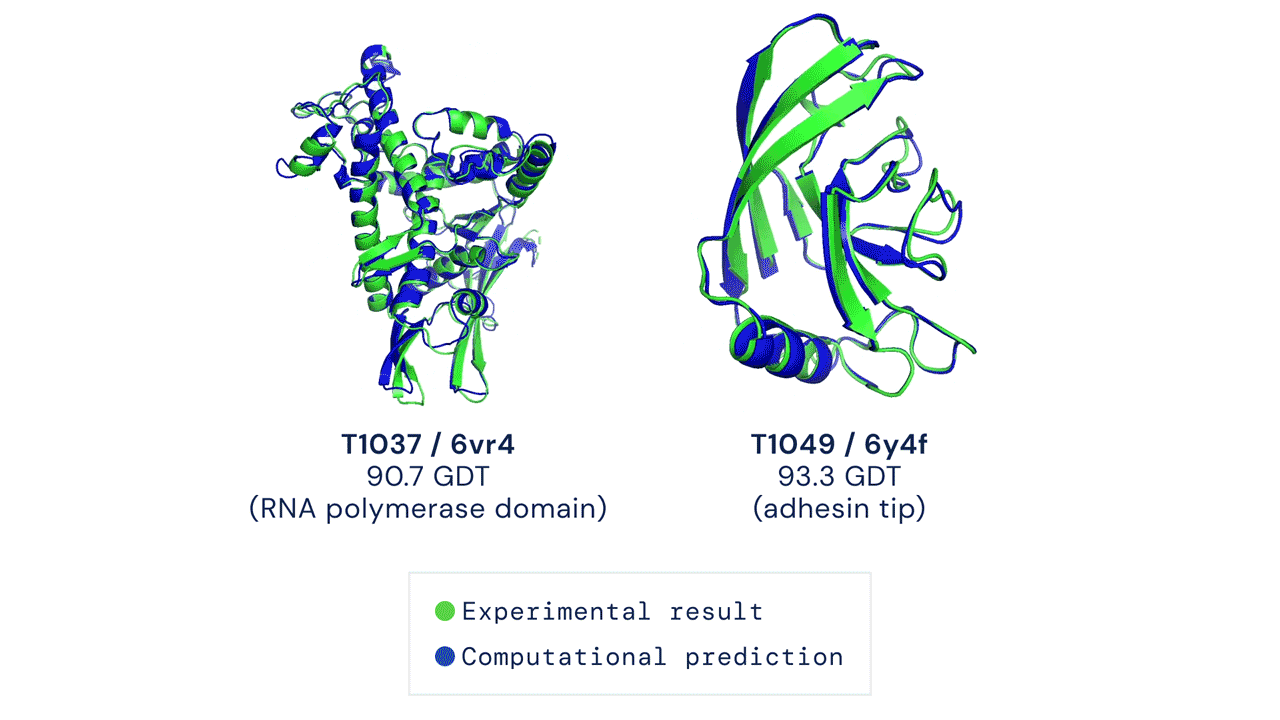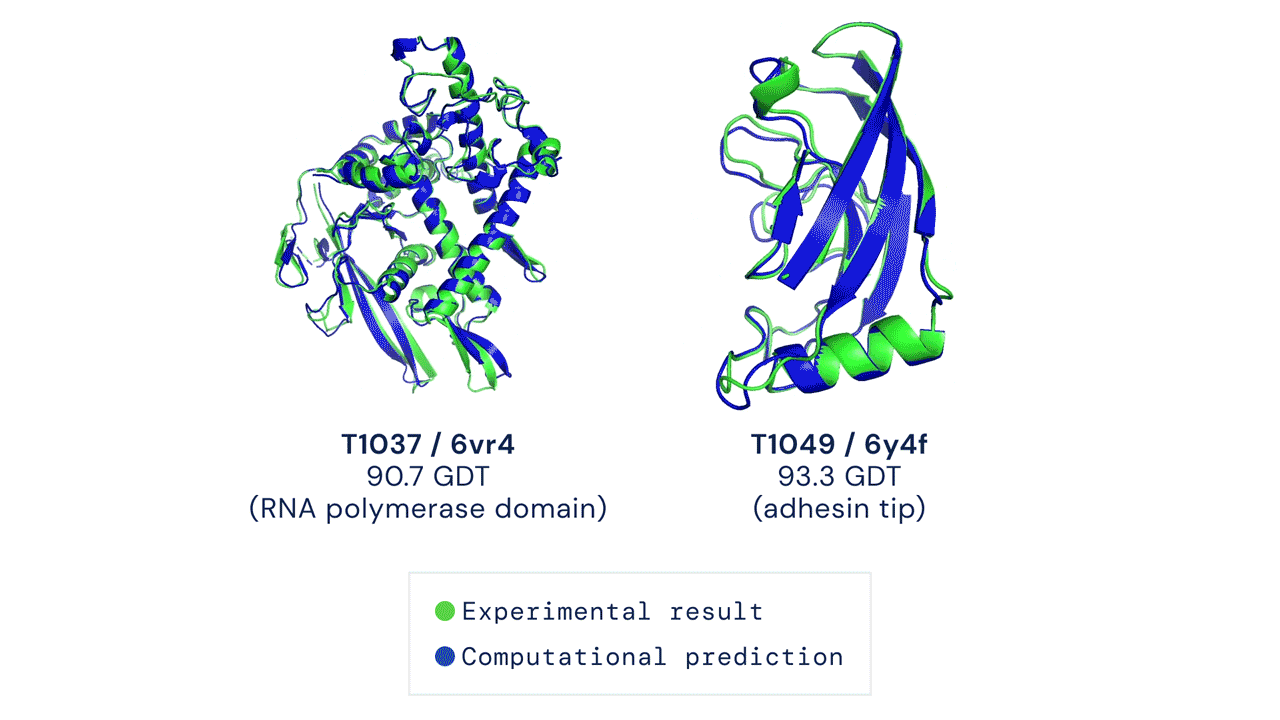
TWO EXAMPLES OF PROTEIN TARGETS IN THE FREE MODELLING CATEGORY. ALPHAFOLD PREDICTS HIGHLY ACCURATE STRUCTURES MEASURED AGAINST EXPERIMENTAL RESULT. Figure from Deepmind
3.1.1 Results of 2020 CASP
A laymen cannot really grasp the significance of this development, so lets listen to what experts say.
An artificial intelligence (AI) network developed by Google AI offshoot DeepMind has made a gargantuan leap in solving one of biology’s grandest challenges — determining a protein’s 3D shape from its amino-acid sequence.
This quote helps to interpret the following numbers.
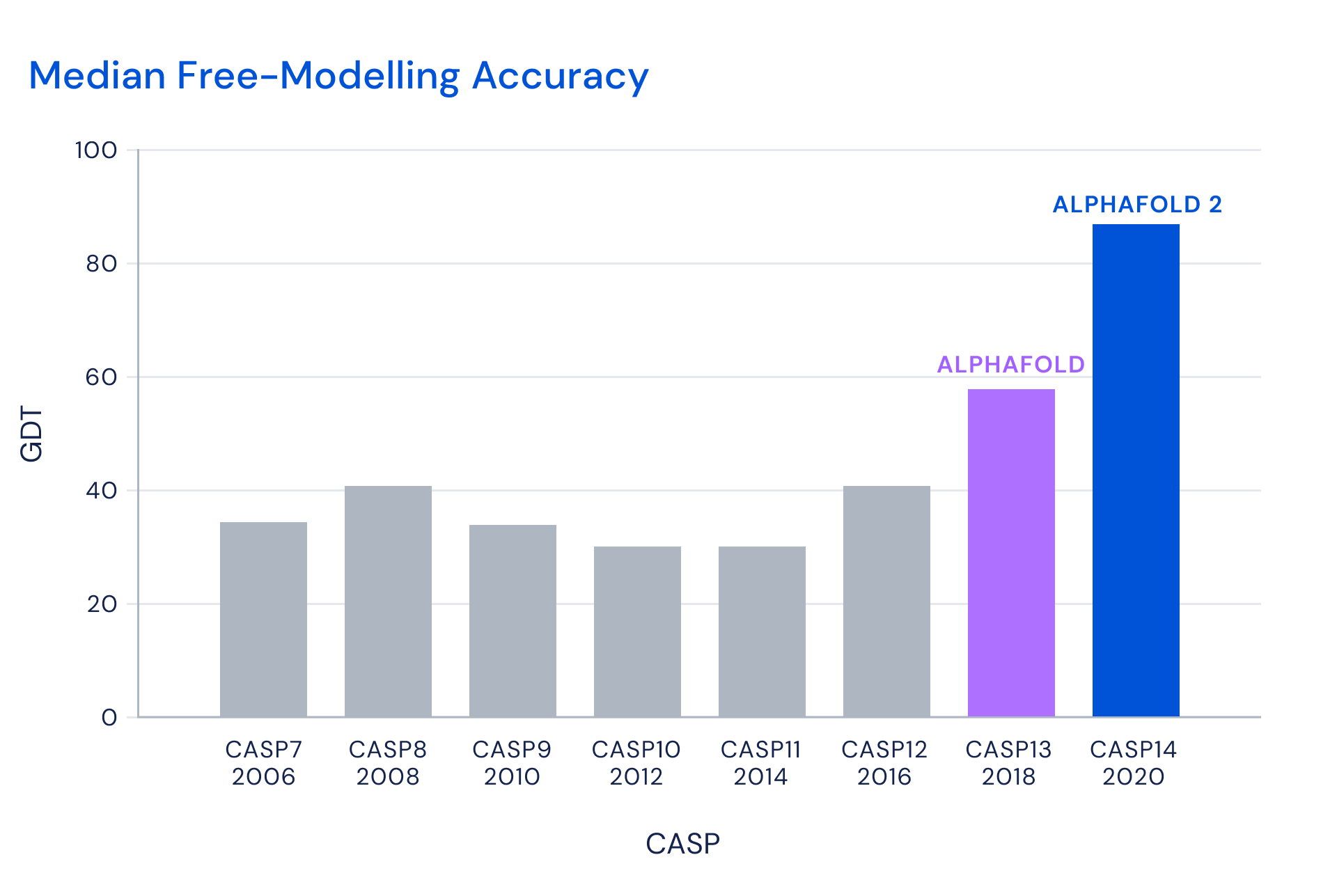
IMPROVEMENTS IN THE MEDIAN ACCURACY OF PREDICTIONS IN THE FREE MODELLING CATEGORY FOR THE BEST TEAM IN EACH CASP, MEASURED AS BEST-OF-5 GDT (Global Distance Test). Figure from Deepmind
Results
- Targets moderate difficult
- AlphaFold ≈ 90 points out of 100
- Other teams around 75
3.1.2 Why does it matter?
The ability to predict the structure of proteins will impact a whole range of industries, there is even talk of creating bacteria which can degrading petroleum products.
Examples of impact
- Life science
- Medicine
- Understand building blocks of cells
- Drug discovery
- quicker
- more advanced
- Environment
- Design bacteria to degrade petroleum products and CO2
- Faster structures
- Example: Structure of a bacterial protein
- Max Planck Institute for Developmental Biology in Tübingen have been trying for years
- X-ray diffraction data
- “The model from group 427 (AlphaFold) gave us our structure in half an hour, after we had spent a decade trying everything” - Lupa, biologist at the Max Planck Institute for
And what is the perspective of a computational biologist at Columbia University?
“I think it’s fair to say this will be very disruptive to the protein-structure-prediction field. I suspect many will leave the field as the core problem has arguably been solved. It’s a breakthrough of the first order, certainly one of the most significant scientific results of my lifetime.
More quotes can be found atscitechdaily.com